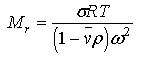Calculating the Molecular Weight
Sednterp can use various methods to calculate molecular weight. First, the program may not calculate the molecular weight at all, but simply use the weight entered by the user. Molecular weight estimates obtained from sequence analysis, gel electrophoresis, gel filtration or other methods may be entered explicitly by pressing the Exact M button and typing in the value. This value will be used in all subsequent calculations. It must be realized that molecular weights calculated by these different means could be in serious error if the composition is based on DNA sequence data and significant post-translational modification of the protein occurs or if the protein exists as an oligomer.
Second, Sednterp will calculate a molecular weight from a sedimentation equilibrium experiment using the formula (a form of equation 4:
Obviously this require that the user enter the temperature, T; sigma, ; the rotor speed (which is entered in RPM and converted to angular velocity squared, ) and that values for and are available. Third, Sednterp will calculate a molecular weight based upon composition. This calculation is performed in the Sample Composition form available by pressing the From Composition button. The minimum molecular weight is estimated by summing up the g/mole of each of the protein's components.
The amino acid composition can be entered as mole-residue per mole-protein in the grid control sorted by one letter or three letter amino acid codes. Entries can also be automatically tabulated from amino acid sequences from a file or pasted in from the clipboard. Entries are provided for the twenty common amino acids, as well as for glx and asx (average of the values for gln and glu and asn and asp respectively) and unk, the average value of the twenty amino acids, for use when the residue identity is unknown (the use of these average values for the amino acids will decrease the accuracy of subsequent calculations). In a separate grid are entries for common ligands/conjugates.
It is important to note that the weights for the amino acids and carbohydrates have been adjusted to reflect the loss of associated with the formation of amide, ester or glycosidic linkages. (The program adds the weight of an additional per polypeptide since there is one less glycosidic linkage than the total number of amino acids.) Other components in the database may not have been corrected in this fashion, so be cautious. (There is an entry for a glycosidic linkage you can use which has a negative molecular weight.) You may browse the values given for the amino acids and ligands in the phyconst.mdb database by choosing Update Databases from the sample form menu. It must be realized that the weight calculated in this fashion can be in serious error if the composition is based on DNA sequence data and significant post-translational modification of the protein occurs or if the protein exists as an oligomer. If the protein is known to be an oligomer, this can be specified on the composition form and all molecular properties will be adjusted appropriately.
Given a composition, other properties of the sample may be calculated including the estimated hydration, the partial specific volume, the charge, the isoelectric point, extinction coefficients, and the extinction spectrum. The results of the charge and absorbance property calculations, and the corresponding graphs, are accessed via the Show Charge and Absorbance Data button on the sample composition form.