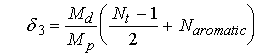Calculating Denatured v-bar Estimates
It is often useful or necessary to measure the molecular weight of proteins in denaturants. In this case, preferential binding of the denaturant to the protein will affect the partial specific volume and must be taken into account. Under these circumstances, vbar must be replaced by the apparent isopotential partial specific volume Φ2 (the nomenclature follows that of Durschlag, where component 2 is the protein). (Ref. 3 6) In general, increasing ionic strength results in a linear increase in Φ2, with the slopes following the Hofmeister series. However, preferential hydration can lead to nonlinear changes in Φ2. Φ2 Values of can be determined as described by Lee and Timasheff (Ref. 42) and then entered directly in using the exact data button.
However, special provisions are made in Sednterp for estimating Φ2 of simple proteins in solvents containing 8 M urea or 6 M guanidinium chloride using described methods. (Ref. 6 37 38 42) In these cases Φ2 determined by equation 9 (ref. 37) replaces vbar in all calculations performed by Sednterp.
Equation 9:
(note that the sign of the second term in this equation is incorrect in ref. 59)
where Φ2 is the calculated value of the isopotential partial specific volume, vbar is from equation 7, ρ is the solvent density, vbar3 is the partial specific volume of the denaturant, δ3 is the number of grams of denaturant bound per gram of protein, δ1 is the hydration in grams of water per gram of protein (described below) and g3 is the number of grams of denaturant per gram of water. For 6 M guanidine HCl at 20 oC, the values are ρ=1.1418 g/ml, vbar3 =0.763 ml/g and g3=1.007 g guanidine-HCl per g H2O.42 For 8 M urea at 20 oC the corresponding values are ρ=1.1152 g/ml, vbar3= 0.763 ml/g and g3 =0.752 g urea per gram of water. (Ref. 37) For both urea and guanidine HCl, is calculated by assuming one molecule of denaturant is bound per pair of peptide bonds and one to each aromatic side chain, including histidine (Ref. 37 42):
Equation 10:
where Nt is the total number of amino acids, Naromatic is the number of aromatic residues, Md is the molecular weight of the denaturant (95.54 for guanidine and 60 for urea) and Mp is the subunit molecular weight for the protein.
Note that erroneous values of Φ2 may be calculated if the overall protein composition has significant levels (> ~10% wt/wt) of non-amino acid constituents. (Ref. 43) However, empirical evidence suggests that reasonable estimates for Φ2 are obtained for glycoproteins in 6 M guanidine HCl by assuming each carbohydrate moiety binds one guanidine molecule and by assuming δ1 is not affected by the presence of the carbohydrate. (Ref. 43) In this case, equation 10 is modified by adding the term [Md/Mp]*Ncarbohydrate to account for the guanidine binding. There has not been sufficient systematic experimental evidence to justify the use of a similar analysis for 8 M urea denaturation of glycoproteins. (Ref. 37)
Non-denaturants may benefit from similar analysis. Timasheff and co-workers have described calculations of Φ2 for simple proteins in solvents containing varying amounts of NaCl, Na2SO4, MgSO4, glycine, -alanine, betaine and CH3COONa. (Ref. 38) It should be noted that such calculations require knowledge of the interaction (i.e. solvation) parameters (δ1 and δ3) for the particular salt, and that it is incorrect to attempt these calculations using interaction data for a different salt. (Ref. 38)